The Interspecies Database compiles anatomical, physiological, and biochemical parameters from various species to help scientists choosing the most appropriate animal species for their research questions. In addition, the database can assist researchers with the refinement, and ultimately also with the replacement, of animal experiments. Users have the flexibility to select various organs, species (and if applicable subspecies, strain or race), age groups, sex, conditions, and additional parameters to facilitate the comparison of values across different species. The database’s filters provide the capability to discern interspecies variations by comparing parameter values among multiple species. The user guide can be downloaded here.
Filter definition
The database includes data from the following Organs: intestine (small, large), gallbladder, kidney, liver, lung, mouth, oesophagus, stomach. Once the user clicks on ‘Choose the organ’ and type an organ of interest, the results will be shown in a drop-down menu to see all the parameters that match with it.
Species
The database includes data from the following Species: dog, human, monkey, mouse, pig, rabbit, and rat (including subspecies and strain or race options where applicable). These species were selected because of their common use in biomedical research.
Age Class
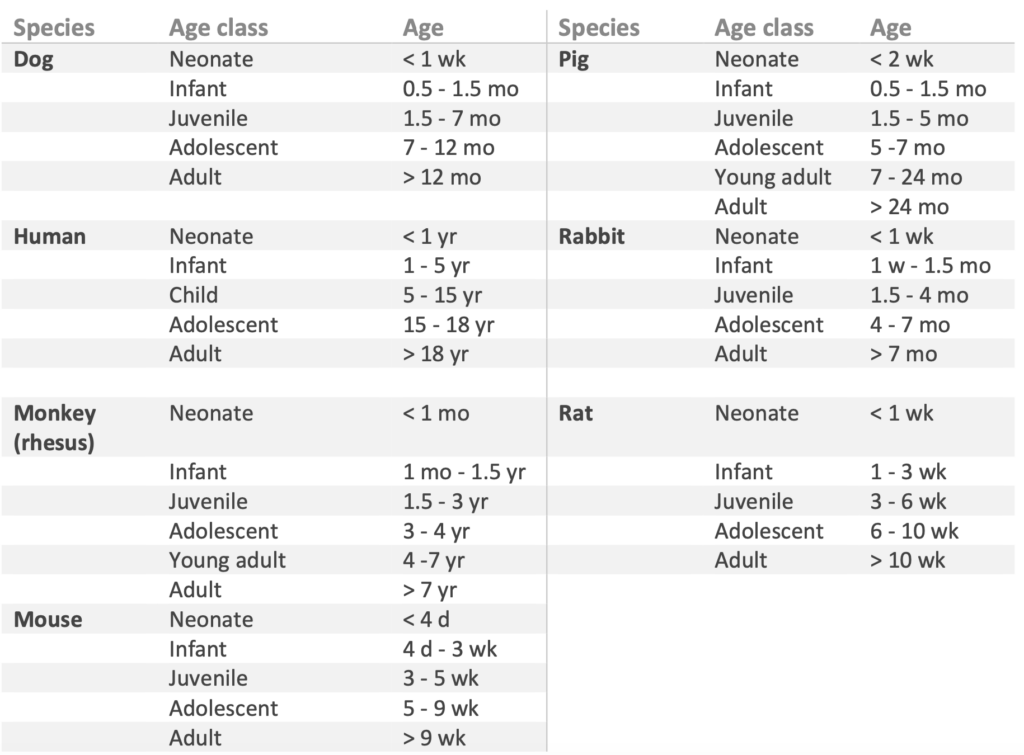
The Age classification is sorted chronologically following this sequence: fetus, neonate, infant, child, juvenile, adolescent, young adult, and adult. The categorisation of age classes is tailoredto the distinct characteristics of the different species included in the database. More detailed information on age class with regard to sub-species and body weight, including the references, can be downloaded here.
Sex
Filter options are F = female, M = male and F + M= both sexes.
Condition
The Condition filter provides options for a diverse range of animals’ conditions such as: pregnancy status, feeding status, PEEP, etc.
Parameters
The Parameter filter contains a variety of extra sub-filters that become visible after clicking on the “+” sign in front of the parameter name. Parameters are organised alphabetically.
The Use of filters and filters options
When no filter is selected, the database shows a screen with default results. The user can start a search by selecting an Organ: only one Organ can be selected at the time. Subsequently, the search can be refined by choosing Species (and if applicable subspecies, strain, or race), Age class, Sex, Conditions, and several additional Parameters. The Species and Parameter filter also contain sub filters that become visible after clicking on the “+” sign in front of name.
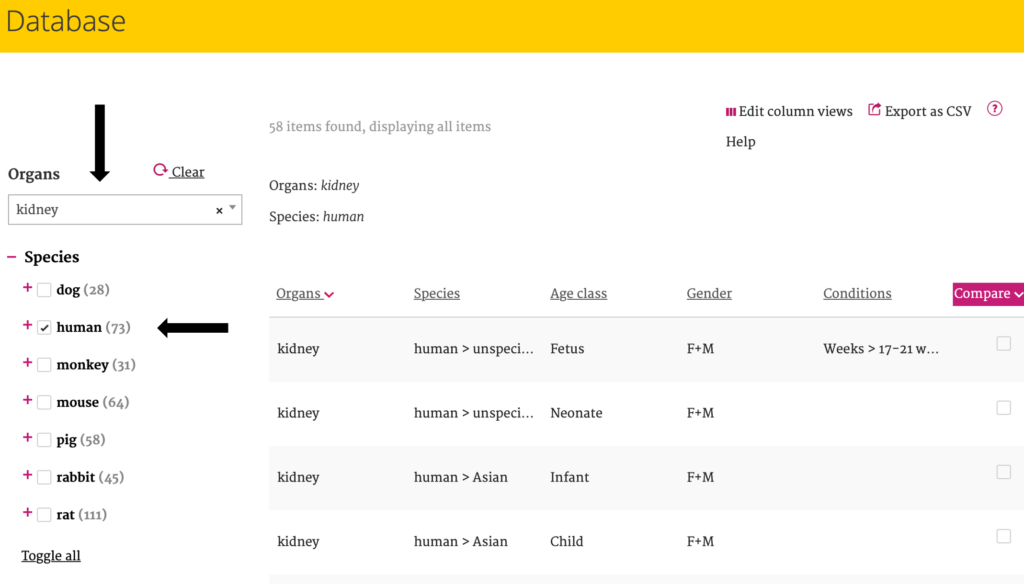
After selecting a filter option, the other available filter options are adjusted to this selection. The chosen filter options are displayed above the data table to keep easy oversight.
For example, when choosing the “Organs – Kidney” and “Species – Human” filter options, only parameters and conditions which are applicable to this selection are shown as further filter options. Behind each filter option, the number of available results is displayed between brackets (for example for “Kidney” there are 73 “Human” records).
Presentation of Results
The results of the Filter selection appear immediately on the screen with the Filter names as column titles in the display. Users can determine which Filters options are visible on the screen by using the ‘Edit Column Views’ option. Clicking on a Column title allows the user to sort the results. Long texts in the results screen are shortened. When moving over the text, the full text appears. By clicking on a result (anywhere), a pop-up window will appear with more detailed information.
In this pop-up window, all data that are available for the selected combination (organ, species, age class, sex, condition, parameters) are shown. For all data, references are available by clicking on the red arrow next to the parameter value. Animals of the same species (or subspecies, strain, or race) can differ on detailed characteristics that have not been added as a filter but that are visible in the pop-up window labelled as ‘Remark’. Such Remarks may provide more information about e.g. breed characteristics but may also provide more detailed information on other parameters.
Comparing results
To compare results, the user can select multiple boxes at the right end of each row and click on ‘Compare’. This action will open the Comparison screen, displaying Filters and Parameters in rows and Species in columns. When the results for a particular parameter are the same across specie(s), the corresponding row is highlighted in green facilitating the identification of similarities and differences in details.
The rows in the Compare screen can be locked, so they will stay in position when scrolling up or down. More detailed information becomes visible by clicking on the Species name at the top. This action triggers a pop-up window similar to the one in the Results screen.
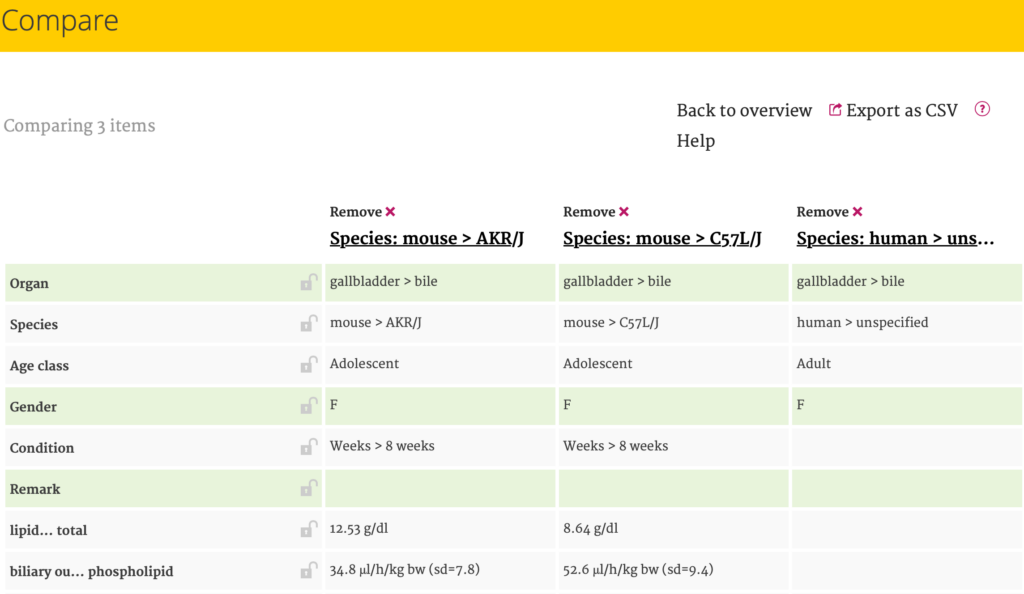
By clicking ‘Back to overview’ the user returns to the Results screen. Removing results can be done by clicking ‘Remove’ in the Compare screen or by untapping (deselecting) the boxes in the Results screen. If this is not done, the results will still appear in the Compare screen after another Selection session (even when ‘clear’ has been clicked) to allow comparison of multiple Selection sessions. Selected results can easily be removed by refreshing the internet browser.
Additional features
The user can edit the display of the columns by clicking the ‘Edit column views’ button on the top right corner of the database. Furthermore, it is possible to export the search results to a CSV-file, by clicking the ‘Export as CSV’ button in the top right corner.
Note: To prevent that all results from differ columns merge into a single column after exporting, users need to use the ‘text to columns’ button to separate the contents of each cell into distinct columns (in Windows Excel).



